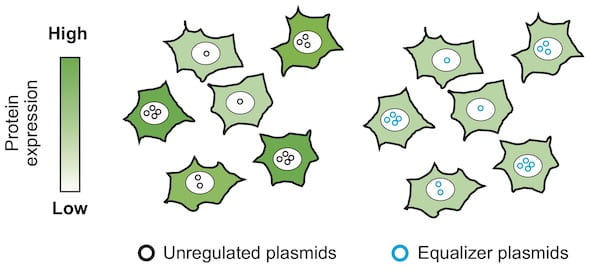
Lab-bred cells like to have it their way, producing recombinant (aka artificial) proteins based on however many plasmids they randomly get. But the intuition and hard work of a Rice University alumnus will go a long way toward preserving the rank and file.
Jin (Harvey) Yang, who graduated from Rice with a degree in bioengineering in 2019, saw a problem he could solve while also working as an intern in a Baylor College of Medicine lab.
His work over two years with Baylor neuroscientist François St-Pierre led to a just-published study in Nature Communications detailing his “Equalizer,” a synthetic circuit that keeps gene expression in colonies of mammalian cells within the bounds of usefulness to the labs studying them.
The project leans heavily on what Yang, now a graduate student at the Massachusetts Institute of Technology, learned under the tutelage of Rice bioengineer Oleg Igoshin, a co-author of the new paper.
“That’s how good Rice undergrads are,” said Igoshin, a professor of bioengineering and biosciences and associate chair of bioengineering. “When you get them in your class, you also learn from them. In this case, Harvey’s effort led to a new collaboration between two labs.”
The Equalizer is an “anti-noise” circuit that assures consistent gene expression from cell to cell, necessary for bioscientists to get accurate experimental results. It combines two subcircuits: a transcriptional negative feedback loop and a post-transcriptional “incoherent” feed-forward control. Together, they minimize variations in protein production.
The two subcircuits don’t normally interact, but Yang showed through both experimentation in the St-Pierre lab and consultation with Igoshin on computational models that the Equalizer could rapidly generate cells lines with minimal variation.
“We didn’t invent the parts, but rather we invented the configuration to build these genetic circuits,” said Yang, in his second year at MIT. “In natural systems, some gene networks must control variation to remain functional and conserve their properties. We were able to use two subcircuits of the gene regulatory network to create the Equalizers to act in a similar manner in the lab.”
Yang took advantage of Rice bioengineering’s allowance for electives and took every course in computational systems biology that Igoshin offered, including those for graduate students. Then he found his opportunity to use what he learned when the Baylor position appeared at a Rice undergraduate research fair.
“There’s a real demand by people who use plasmids — which, basically, every lab does — to express proteins,” he said. “The issue is that after you introduce the plasmid into individual cells, the number of plasmid copies will differ among cells. Because of this variation, the resultant protein expression level will differ a lot among the cells.
“That’s why we thought it would be great if we could have a genetic circuit able to buffer the difference in plasmid copies,” he said.
Yang thought one negative feedback circuit would do the trick. “But I tested it in the lab, and it didn’t work that great.”
That’s where his concurrent systems biology training with Igoshin paid off.
“This is similar to how engineers work; they draw up their plans, create a model and then build their structure,” said Igoshin, who also teaches as part of Rice’s Systems, Synthetic and Physical Biology program. “In this case we were able to show the effectiveness through computational models before it was then synthetically engineered in the lab.”
Completing the work during the pandemic was no small task, according to Yang, who had to sort out visa issues while anticipating and then diving into his graduate studies at MIT. “The framework of the paper was already established before I left Rice, but I spent my whole summer the year I graduated working on it,” he said. “We submitted the paper and the reviewers had some suggestions that led to additional modeling and experiments.”
At MIT, Yang continues to split his work between modeling and experimenting, this time focusing on how the 3D organization of the genome in cells regulates gene expression.
“I was pretty lucky to have Oleg’s mentorship on the computational side,” he said. “I was able to dig deep into building models and making predictions, and it’s quite valuable in minimizing experimental work.”
Co-authors of the paper are Rice graduate student Jihwan Lee and Baylor postdoctoral associate Michelle Land and research assistant Shujuan Lai.
The research was supported by the Welch Foundation, a Klingenstein-Simons Fellowship Award in Neuroscience, the McNair Medical Foundation, the National Science Foundation (including the NSF-supported Center for Theoretical Biological Physics at Rice), the National Institutes of Health, and the Cancer Prevention and Research Institute of Texas.