Rice theorists show how random processes cancel out to ensure microbial health
Fat bacteria? Skinny bacteria? From our perspective on high, they all seem to be about the same size. In fact, they are.
Precisely why has been an open question, according to Rice University chemist Anatoly Kolomeisky, who now has a theory.
A primal mechanism in bacteria that keeps them in their personal Goldilocks zones -- that is, just right -- appears to depend on two random means of regulation, growth and division, that cancel each other out. The same mechanism may give researchers a new perspective on disease, including cancer.
The “minimal model” by Kolomeisky, Rice postdoctoral researcher and lead author Hamid Teimouri and Rupsha Mukherjee, a former research assistant at Rice now at the Indian Institute of Technology Gandhinagar, appears in the American Chemical Society’s Journal of Physical Chemistry Letters.
“Everywhere we see bacteria, they more or less have the same sizes and shapes,” Kolomeisky said. “It’s the same for the cells in our tissues. This is a signature of homeostasis, where a system tries to have physiological parameters that are almost the same, like body temperature or our blood pressure or the sugar level in our blood.
“Nature likes to have these parameters in a very narrow range so that living systems can work the most efficiently,” he said. “Deviations from these parameters are a signature of disease.”
Bacteria are models of homeostasis, sticking to a narrow distribution of sizes and shape. “But the explanations we have so far are not good,” Kolomeisky said. “As we know, science does not like magic. But something like magic -- thresholds -- is proposed to explain it.”
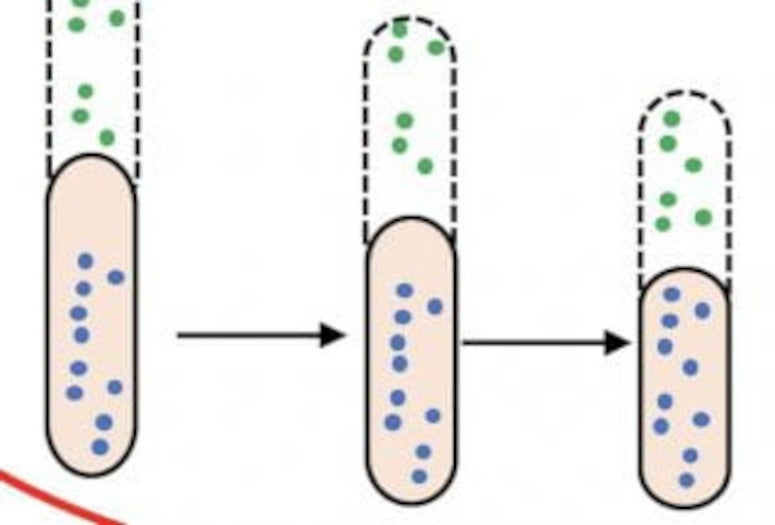
For bacteria, he said, there is no threshold. “Essentially, there’s no need for one,” he said. “There are a lot of underlying biochemical processes, but they can be roughly divided into two stochastic chemical processes: growth and division. Both are random, so our problem was to explain why these random phenomenon lead to a very deterministic outcome.”
The Rice lab specializes in theoretical modeling that explains biological phenomena including genome editing, antibiotic resistance and cancer proliferation. Teimouri said the highly efficient chemical coupling between growth and division in bacteria was far easier to model.
“We assumed that, at typical proliferation conditions, the number of division and growth protein precursors are always proportional to the cell size,” he said.
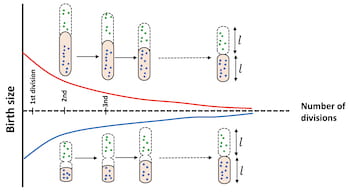
The model predicts when bacteria will divide, allowing them to optimize their function. The researchers said it agrees nicely with experimental observations and noted manipulating the formula to knock bacteria out of homeostasis proved their point. Increasing the theoretical length of post-division bacteria, they said, simply leads to faster rates of division, keeping their sizes in check.
“For short lengths, growth dominates, again keeping the bacteria to the right size,” Kolomeisky said.


The same theory doesn’t necessarily apply to larger organisms, he said. “We know that in humans, there are many other biochemical pathways that might regulate homeostasis, so the problem is more complex.”
However, the work may give researchers new perspective on the proliferation of diseased cells and the mechanism that forces, for instance, cancer cells to take on different shapes and sizes.
“One of the ways to determine cancer is to see a deviation from the norm,” Kolomeisky said. “Is there a mutation that leads to faster growth or faster division of cells? This mechanism that helps maintain the sizes and shapes of bacteria may help us understand what’s happening there as well.”
Kolomeisky is a professor of chemistry and of chemical and biomolecular engineering. The Welch Foundation, the National Science Foundation and Rice’s Center for Theoretical Biological Physics supported the research.