In your cells, it’s almost always spring. Or at least springy.
Bioscientists have known for some time that chromosomes tend to express their protein products in bursts, rather than in a steady manner. A new theoretical study by Rice University scientists, detailed in the Biophysical Journal, aims to better explain the process that combines chemical reactions and mechanical forces.
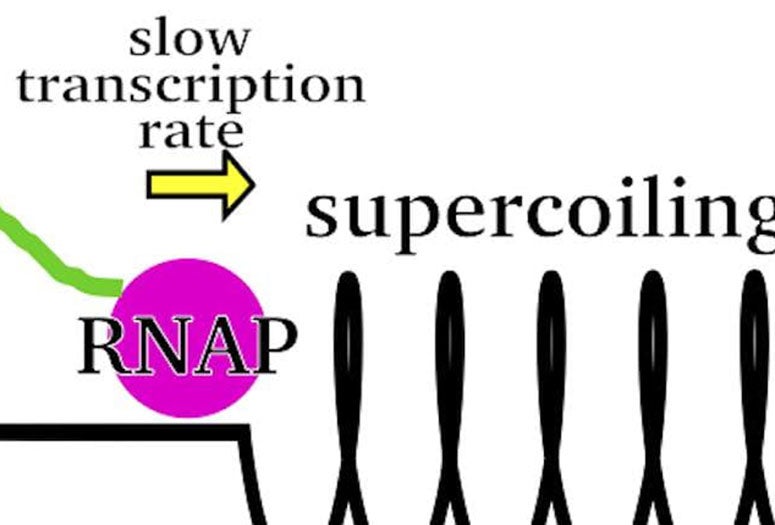
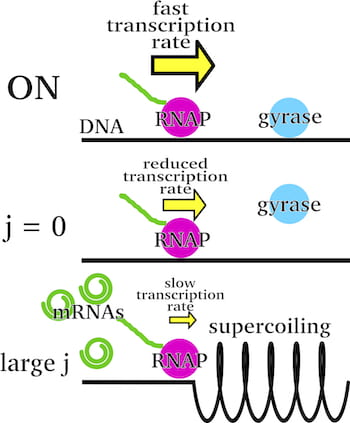
Rice chemist Anatoly Kolomeisky and applied physics graduate student and lead author Alena Klindziuk built the first simplified analytical model of “bursting” to show how pressure from a RNA polymerase enzyme triggers the rush of RNA production, but only to the degree that it can push a coil of DNA.
As it compresses like a spring, that DNA “supercoil” continues to express RNA — which goes on to make the proteins themselves — until the enzyme can push no more. It isn’t until another enzyme, a gyrase, comes along to release the tension that production can start anew.
“With advances in experimental techniques, people are able to measure how much RNA you are producing, and so it was a naive expectation that the speed of production was more or less constant,” said Kolomeisky, a chemist by title whose group has long been interested in how biochemical reactions work to power biological mechanisms, and vice versa.
“It was surprising when we found it actually doesn't work this way,” he said. “A lot of RNA is produced and then there's a period of silence. RNA is produced in a very bursty behavior, but the molecular details have been lacking.”
He said the way RNA polymerase aligns with the double-helical DNA coils it in the process. “It rotates, putting mechanical constraints on DNA,” Kolomeisky said. “A spring is an excellent example. The more you push a spring, the harder it becomes to push.
“We think the RNA polymerase coils DNA to start RNA production,” he said. “At the beginning of the process, you get a burst, but the process slows down as it squeezes the spring. Then gyrases come in; they untangle this supercoil so that normal production can begin again.” At the same time, he said gyrases also relieve negative stress created on the other side of the polymerase.
“Essentially, we created the first quantitative energetic model that explains this burstiness,” Kolomeisky said. “We are able to interpret the experimental data (gathered in experiments on bacteria) and found this supercoil exists.”
He said the calculations show the DNA “spring” is relatively weak. “That has biological significance because it means we can more easily regulate the process by regulating gyrases,” he said.
Klindziuk noted there are many other players in the process that ultimately need to be accounted for. “We could have added many effects, like transcriptional and other epigenetic factors,” she said. “We want to make a model where there are multiple polymerases on DNA. In this model, we only had one, but in reality, there are many polymerases. There might be effects from polymerase traffic, like they're bumping into each other and stopping and resuming their activity.”
“This is an example of advanced experimentation that led us to seek a significant theoretical solution,” Kolomeisky said. “It usually happens in the opposite direction, but this time experiments were able to visualize the process, and that led us to think about and start to explain it.”
Visiting undergraduate student Billie Meadowcroft is a co-author of the paper. Kolomeisky is a professor of chemistry and of chemical and biomolecular engineering and chair of Rice’s Department of Chemistry.
The Welch Foundation, the National Science Foundation and Rice’s Center for Theoretical Biological Physics supported the research.