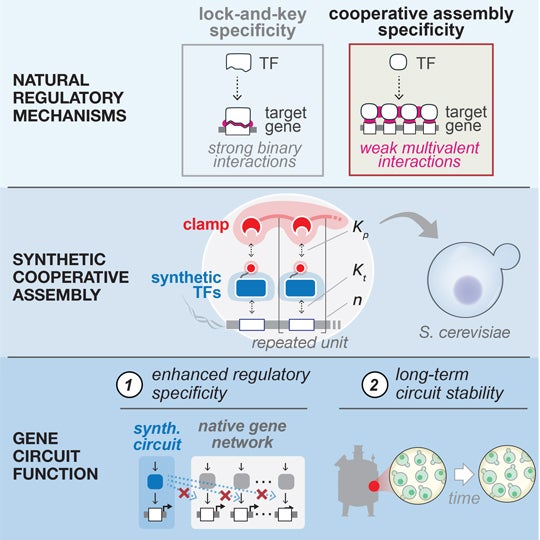
Bioengineers can tailor the genomes of cells to create “cellular therapies” that fight disease, but they have found it difficult to design specialized activating proteins called transcription factors that can throw the switch on bioengineered genes without occasionally turning on some of the cell’s naturally occurring genes.

In a study published online in Cell, bioengineers from Rice University, Boston University, Harvard Medical School, Dartmouth College and Harvard University’s Wyss Institute showed they could all but eliminate such “off-target” gene activations using a method that takes its cue from nature.
“We made our transcription factors weaker,” said co-senior author Caleb Bashor, an assistant professor of both bioengineering and biosciences at Rice who helped lead the study. “Because they bind more weakly overall, the chances of their binding off-target drop to almost nothing.”
In general, bioengineers have leaned toward designing strong-binding transcription factors to help ensure target genes get activated when they are supposed to. While weakening transcription factors seems counterintuitive, Bashor’s research group has collaborated with the group of BU’s Ahmad “Mo” Khalil for years to build and test tools that employ weaker transcription factors by making them work in teams.
“Transcription factors act as the ‘wiring’ in gene circuits by linking together the expression of different genes in the circuit,” Bashor said.
Gene circuits are sets of genes that work together to perform a specific function. For example, in previous work Bashor and colleagues implemented circuits that performed programmable Boolean logic, signal processing, analog-to-digital conversion and other complex tasks.
Each transcription factor activates its specific target gene by binding with a particular sequence of DNA that activates that gene. Bioengineers can use one transcription factor to turn a particular element of a gene circuit on, for example, another to turn its output from low to high and yet another to turn it off.
To ensure that their weakened transcription factors activated their target genes when called upon, Bashor, Khalil and colleagues employed a phenomenon called cooperative assembly. In their cells, a transcription factor only activates its target by first merging with one or more other transcription factors to form a large protein complex. The assembled complex, behaving as a single unit, activates the target gene.
“Our design makes them strong as a group, but weak alone,” Bashor said. “It ensures that the only genes they can get together to activate are the ones in the circuit. The result of this is gene circuits that work normally but are also ‘stabilized’ and remain in the cell long term.”
Bashor said nature’s use of a similar strategy in humans and in other complex life was an inspiration for the project.
To illustrate the work’s potential, he used the example of cell-based therapies. In many, the number of engineered cells a patient receives is far fewer than the number needed to produce a therapeutic effect. Meaning, the treatment is only effective if the engineered cells thrive, reproduce and grow into a population that’s large enough to take on the disease.
“Any burden imposed through off-target interactions lowers the odds of success,” he said. “Our approach offers a generalizable set of rules engineers can use to insulate gene circuits from the host cell and mitigate off-target burdens.”
The research was funded by the National Institutes of Health (EB029483, EB027793), the National Science Foundation (2027045, 1921677), the Office of Naval Research (N00014-21-1-4006) and a Department of Defense Vannevar Bush Faculty Fellowship (N00014-20-1-2825).
- Peer-reviewed paper
-
“Cooperative assembly confers regulatory specificity and long-term genetic circuit stability” | Cell | DOI: 10.1016/j.cell.2023.07.012
Authors: Meghan D. J. Bragdon, Nikit Patel, James Chuang, Ethan Levien, Caleb J. Bashor and Ahmad S. Khalil
- Image downloads
-
https://news-network.rice.edu/news/files/2023/08/0814_ASSEMBLY-fg1-lg.jpg
CAPTION: Bioengineers can control cell behavior with artificial gene circuits, but it has proven difficult to design specialized proteins called transcription factors (TF) that can activate synthetic genes without occasionally activating some naturally occurring genes. Rice University bioengineer Caleb Bashor co-led the development of a generalizable method engineers can use to avoid “off-target” gene activation by creating transcription factors that bind weakly with DNA when they are alone and strongly (top right) when they cooperatively assemble (center) with other transcription factors. In experiments, the researchers showed cooperative assembly could improve the long-term stability of synthetic gene circuits (bottom right) by eliminating ill effects of off-target binding (bottom left). (Illustration courtesy of Caleb Bashor/Rice University)https://news-network.rice.edu/news/files/2023/08/0814_ASSEMBLY-cb28-lg.jpg
CAPTION: Caleb Bashor is an assistant professor of both bioengineering and biosciences at Rice University. (Photo by Jeff Fitlow/Rice University) - Related stories
-
Reporters broadcast live, on-the-scene, inside living cells – July 10, 2023
https://news.rice.edu/news/2023/reporters-broadcast-live-scene-inside-living-cellsBashor wins NIH grant to engineer artificial protein signaling networks – Aug. 3, 2021
https://bioengineering.rice.edu/news/bashor-awarded-nih-grant-engineer-artificial-protein-signaling-networks-within-cellsBioengineers program cells as digital signal processors – April 18, 2019
https://news2.rice.edu/2019/04/18/bioengineers-program-cells-as-digital-signal-processors-2/ - Links
-
Bashor lab: bashorlab.rice.edu/
Rice Department of Bioengineering: bioengineering.rice.edu/
Rice’s George R. Brown School of Engineering: engineering.rice.edu/
- About Rice
-
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation’s top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 4,240 undergraduates and 3,972 graduate students, Rice’s undergraduate student-to-faculty ratio is just under 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for lots of race/class interaction and No. 4 for quality of life by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger’s Personal Finance.

