NIH backs Rice University study of delay in gene transcription networks
A Rice University researcher and his colleagues have received a National Institutes of Health grant to study how delays in gene transcription – life’s most basic messaging system – affect cellular processes.
Matthew Bennett, an assistant professor of biochemistry and cell biology, will receive more than $1 million over five years to gather and combine data from computer simulations and live cells. The goal is to create techniques to generate and analyze models of gene networks that incorporate delay as part of their process.
“Delay in transcriptional signaling is an unavoidable consequence of the way biochemistry works,” said Bennett, a theoretical physicist by training who turned to synthetic biology as a postdoctoral researcher before joining Rice in 2009. “Once a gene is activated, often as a response to a molecule being introduced into the cell, it takes time for the results to come to fruition. Eventually, the DNA must be transcribed into RNA, and the RNA must be translated into protein. Then, sometimes, the protein has to be modified or has to fold.”
Bennett and his team want to know precisely what happens and why in that cascade of events, which can take minutes. “We want to be able to create accurate mathematical models of gene networks to predict how they function and how they fail, so we can design new synthetic networks and know what they’re going to do before we build them,” he said.
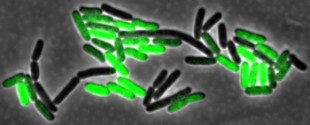
E. coli bacteria, shown growing in a microfluidic device, will be one focus of a Rice University study to understand how delays in gene transcription affect cellular processes. Microfluidic devices allow researchers to track how gene expression patterns of individual cells change over time. Image by Bennett Lab/Rice University
Synthetic biology has become an increasingly hot topic as researchers create biological systems not found in nature. (A leader in the field, J. Craig Venter, will speak at Rice Oct. 10 as part of the university’s Centennial Lecture Series.)
Bennett sees the process as similar to sophisticated electronic design, in which the genetic equivalent of logic gates can be used to program circuit-like behavior in living things. The resulting genomes can be used in cells for information processing, materials fabrication, chemical sensing, the production of energy and even as food.
“We’ve learned that delays can have a significant impact on the function of genetic networks, and the discoveries have changed our fundamental understanding of how genes talk to one another,” Bennett said. “There are many processes in cells that require specific timing in order to operate: stress responses, circadian oscillations, cellular growth and division. We’ve found the dynamics of these networks are important to their function, and delay can have a profound effect. Understanding that delay and being able to model it is critical for accurate computational simulations.”
While simple models are able to help build simple cellular “circuits,” Bennett felt a more sophisticated model will expand the possibilities for synthetic biology and lower the cost of engineering new synthetic microbes.
Much of the modeling will take place at the University of Houston, where co-investigators Krešimir Josic is an associate professor and William Ott is an assistant professor of mathematics; the synthesis of gene networks in bacteria will happen at Bennett’s Rice lab.
Bennett also noted the new work will call for an interdisciplinary approach that draws upon expertise from microfluidic engineers and theoretical biologists. In the big picture, he hopes better computational models enhance understanding of living systems.



Leave a Reply